GalaxyPepDesign
Predicted motif-based peptide design using conformational space annealing
Method for GalaxyPepDesign
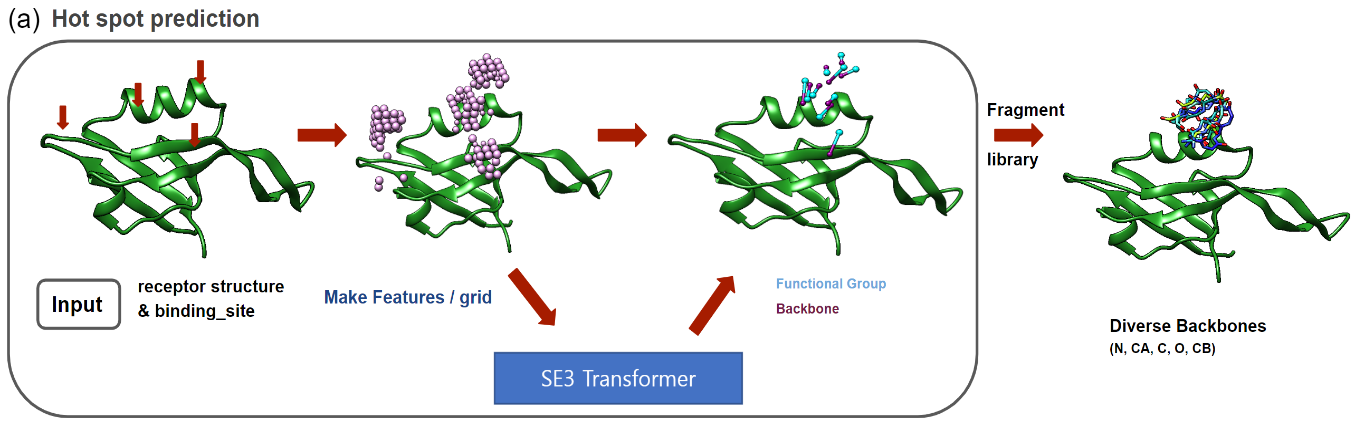
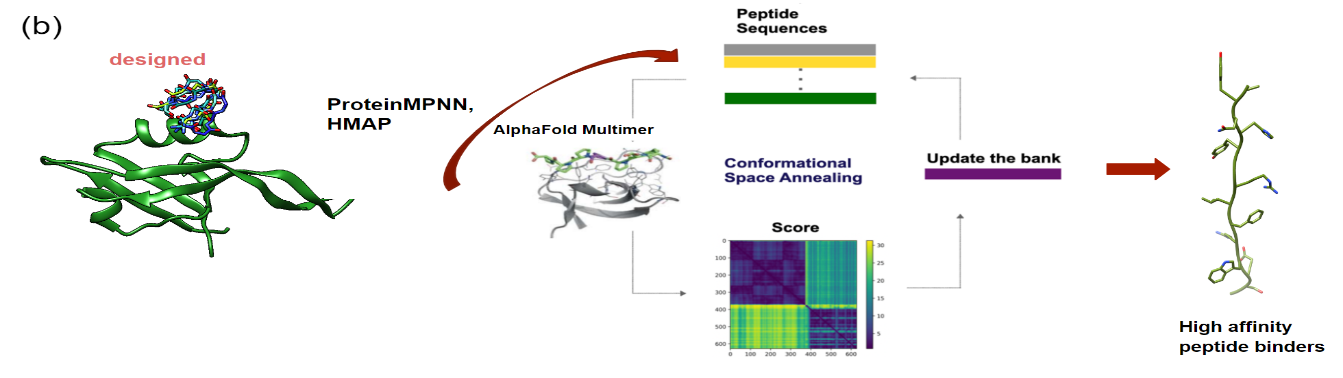
GalaxyPepDesign generates peptide binders from input protein structure and binding site residues by combining information of binding motifs predicted by MotifNet, protein complex prediction with AlphaFold-Multimer and AlphaFold-Multimer confidence score based optimization by Conformational Space annealing.
Inputs for GalaxyPepDesign
First, you have to enter the following information required for design
- Job name: Enter job name for your recognition.
- E-mail address: GalaxyWEB server will send progress reports of your job (if provided).
- Receptor structure: You have to provide a receptor(protein) structure in the PDB format.
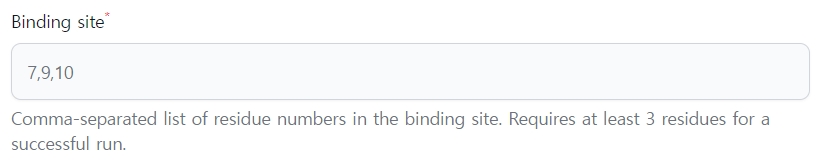
- Binding site: You have to provide probable interface residues at receptor structure as Comma-separated list. Requires at least 3 residues for a successful run.
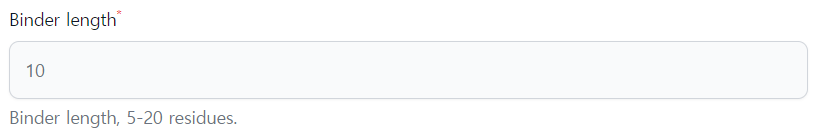
- Binder length: You have to provide the length of the peptide to be designed. The range of binder length should be 5-20 residues.
Outputs for GalaxyPepDesign
Designed peptide sequences
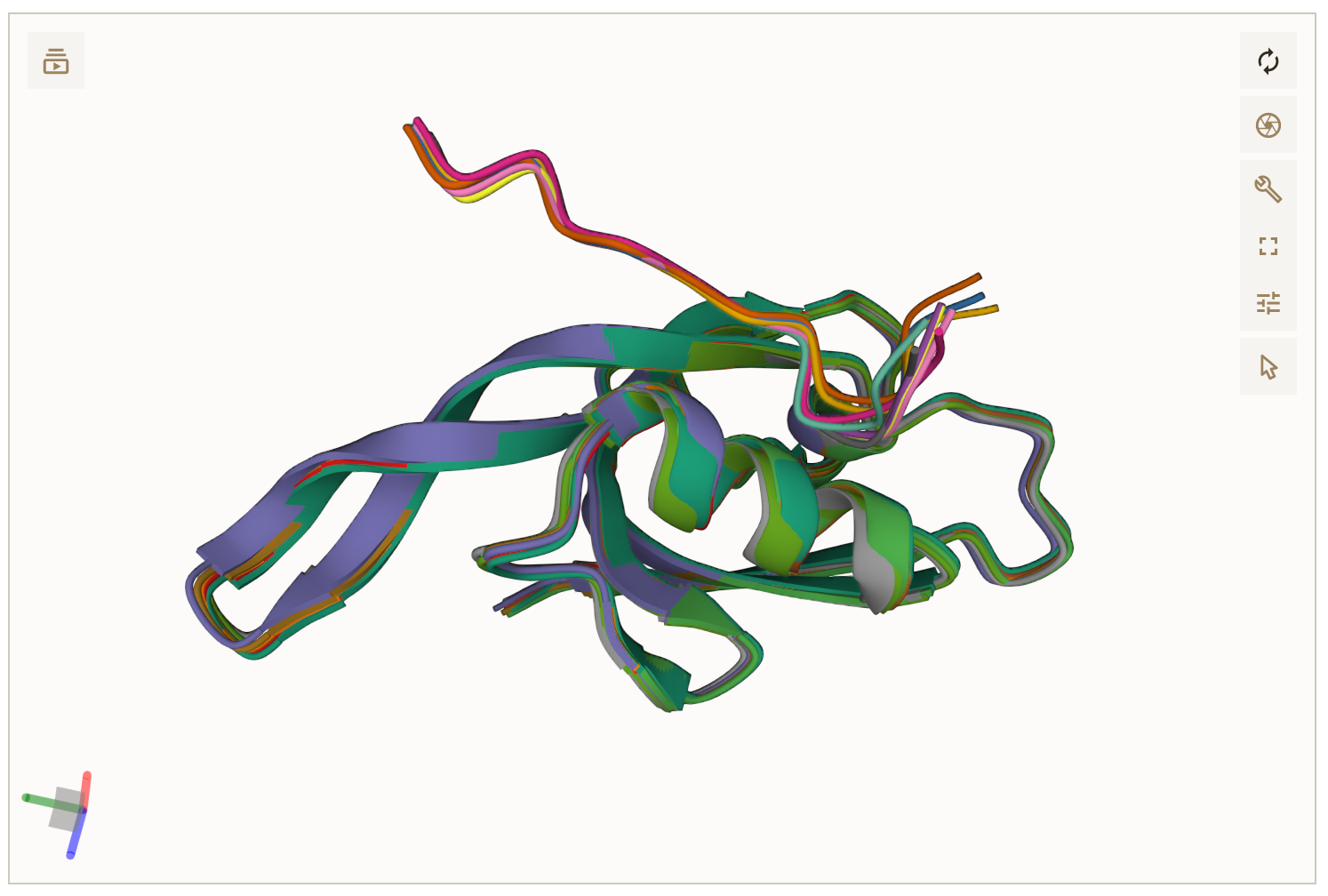
Up to 10 Designed peptide sequences and protein-peptide complex structures are shown.

Predicted structure (AlphaFold-Multimer)
Predicted AlphaFold_Multimer protein-peptide complex model structures are provided in PDB format.
Download
Users can download the output (results.tar.gz) containing peptide sequences (output.fa) and model structures (receptor_{1..10}.pdb) in the 'Downloads' section.