AlphaFold2 Modeling for GPCR
Sumin Lee, Seeun Kim, Chaok Seok* and Hahnbeom Park*
To predict GPCR-ligand complex structures, use AlphaFold-template biasing for receptor and flexible docking tool like GALD. For peptide docking, use AlphaFold-multimer with recycle number 10.
Receptor modeling and small molecule docking for GPCR
This protocol assumes that the user knows the target protein's activation state and the binding site. If this information is unavailable, the user can infer the activation state based on the ligand's type(agonist, antagonist) and use a binding site prediction program like GalaxySite1. Due to the significant conformational change of GPCR upon activation, receptor modeling according to activation states and side-chain flexible docking tools are preferred to get more accurate pose predictions.
- Run multistate AlphaFold by ref. 22 with template biasing option 'active' or 'inactive.' This option constrains the template database to proper activation-form GPCR templates.
- Run a flexible docking tool like GALigandDock3
Examples
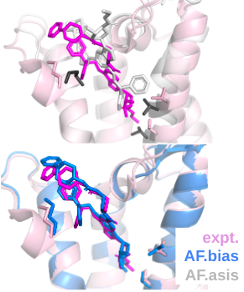
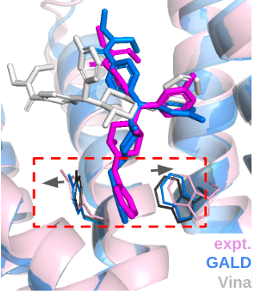
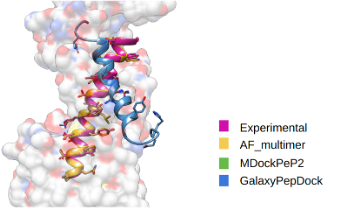
Peptide docking with AF multimer for GPCR
AlphaFold multimer predicts not only protein complex but also protein-peptide complex well. It is known that more recycling (feeding the predicted structure as a template to the evoformer) could improve the final structure, but it takes longer. We recommend setting recycle number as 10.
- Run AlphaFold multimer with
- receptor sequence, and
- peptide sequence.
Examples
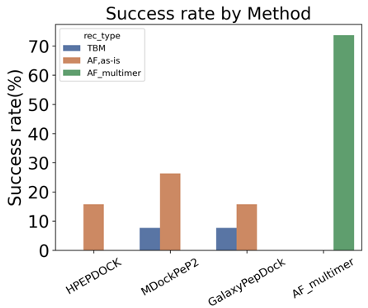
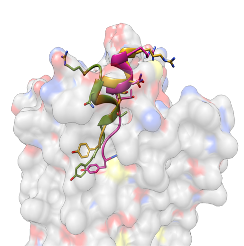
-
Heo, L.; Shin, W. -H.; Lee, M. S.; Seok, C. GalaxySite: ligand-binding-site prediction by using molecular docking, Nucleic Acids Res. 2014, 42 (W1), W210-W214. DOI GalaxyWEB ↩
-
Heo, L.; Feig, M. Multi-State Modeling of G-protein Coupled Receptors at Experimental Accuracy, Proteins 2022, 90 (11), 1873-1885. DOI GitHub ↩
-
Park, H. et al. Force field optimization guided by small molecule crystal lattice data enables consistent sub-angstrom protein–ligand docking, J. Chem. Theory Comput. 2021, 17 (3), 2000-2010. DOI Homepage ↩